This post was originally published on here
A team of researchers has generated one of the most detailed 3D maps of human chromosomes to date, cataloging over 140,000 DNA looping interactions to reveal how genetic material is folded and organized within the cell nucleus.
The findings, published in the journal Nature, mark a significant advancement for the 4D Nucleome Project, an initiative dedicated to understanding the three-dimensional organization of chromosomes and how it changes over time.
By charting these complex architectures, scientists hope to better understand how genetic variations, including those linked to disease, reshape the structure and function of the genome.
Researchers focused on mapping the 3D DNA of human embryonic stem cells
Contrary to the static X-shaped illustrations found in biology textbooks, chromosomes are dynamic, thread-like structures that fold into specific three-dimensional forms. This folding is not merely structural; the arrangement and placement of these forms play crucial roles in maintaining cellular functions, regulating gene expression, and facilitating DNA replication.
To map this architecture, the research team focused on two human cell types: H1 embryonic stem cells and immortalized foreskin fibroblasts. Through a multi-step approach involving various genomic assays, they identified 141,365 regulatory looping interactions in the stem cells and 146,140 in the fibroblasts.
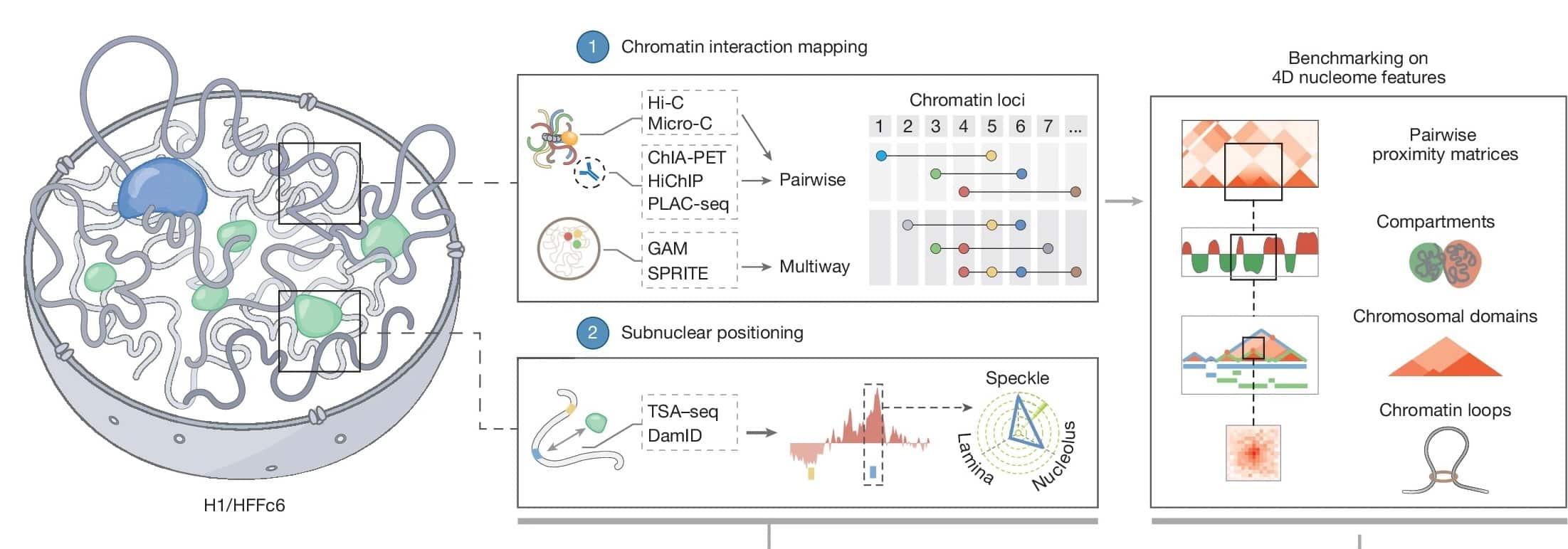
These structures, known as chromatin loops, are fundamental to the genome’s organization in 3D space. By bringing separate regions of DNA into contact, the loops promote the interactions required to switch genes on or off, a layer of complexity that the original Human Genome Project, which listed the DNA sequence but not its architecture, could not fully explain.
Researchers used IGM to create single-cell models
The study utilized an Integrative Genome Modeling (IGM) platform, which synthesized data from laboratory tests to generate 1,000 distinct 3D models of the genome for individual cells.
Crucially, the team also presented computational methods that use deep learning to predict genome folding based solely on the DNA sequence. This development allows researchers to create single-cell models that visualize how genes interact with distant regulatory elements throughout three-dimensional space.

The authors highlight that these models could reveal the specific DNA sequences that guide genome folding. Continued exploration could provide clinicians with key insights into how specific genetic disorders alter the physical landscape of the nucleus.